Presenter:
Luvena Ong
Abstract:
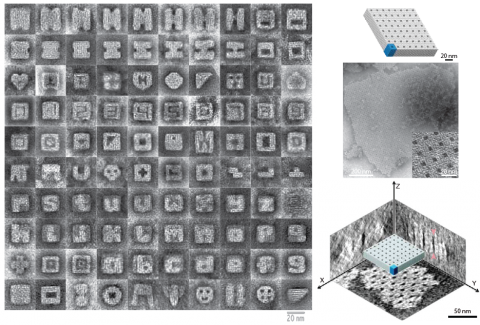
Nanoscale assemblies are useful for arranging and orienting different functional moieties. A number of approaches have been developed to obtain these nanoscale features and to engineer unique material properties. Self-assembly of biopolymers is an attractive method of fabricating nanoscale features, because the structural information is encoded in the sequence of monomers. Nucleic acid structures are easy to program because of the predictable Watson-Crick base-pairing. Discrete 2D and 3D structures, periodic and algorithmic 2D lattices, extended ribbons and tubes, 3D crystals, and polyhedra have been assembled using DNA. Generally, these structures are assembled using modular tiling methods or DNA origami; however, the tiling methods often result in structures less complex than those achievable by origami methods. To assemble complex structures in a modular manner, we selected subsets of single-stranded DNA “bricks” from a library of strands to construct over 100 different discrete DNA shapes. We also extended this assembly method to form complex, depth-defined crystals by designing strands with complementary segments in opposing faces of the unit structure. These crystals can grow to micron size and can be designed with different cavities or tunnels. Keywords: DNA Nanotechnology, Self-assembly, Three-dimensional patterning